If you are not acquainted with R programming or you want to look at simple examples, you can run the GenometriCorr Wizard
|
|
| Subscribe to GenometriCorr-users |
| Visit this group |
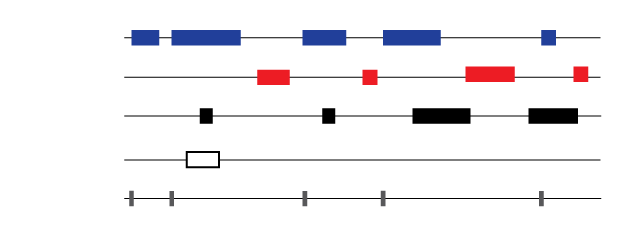
Genetic data are generally arranged spatially along chromosomes, and in many genetic datasets, each point has a numerically defined location in the genome, and spatial proximity often correlates with functional similarity. When comparing two genetic datasets, we can take advantage of spatial measures to test whether genetic datasets are independent of each other with regards to position on the genome, or whether they are situated in a mutually nonrandom way.
The page is the home of our R package GenometriCorr that performs several carefully chosen spatial tests of independence on genome-wide data. It is described in the PLOS Comp Bio publication.
|
If you are not acquainted with R programming or you want to look at simple examples, you can run the GenometriCorr Wizard |
|
|||